Antimicrobial Compounds and the Gastrointestinal Microbiome: Implications for Health and Disease
Author
Amanda Buerger
Ph.D. Candidate, Department of Environmental and Global Health,
College of Public Health and Health Professions, University of Florida
The number of bacterial cells in the human body is estimated to be equal to the number of human cells (Sender et al. 2016). Many bacteria live in the gastrointestinal system, constituting a functional bacterial microbiome, or an ecosystem of bacteria. Evidence is mounting that these bacterial communities play a large role in the health of humans, both in the gastrointestinal system and in other organ systems. A One Health approach will be beneficial to further understand the function of the microbiome and explore how environmental perturbations affect the microbiome and human health.
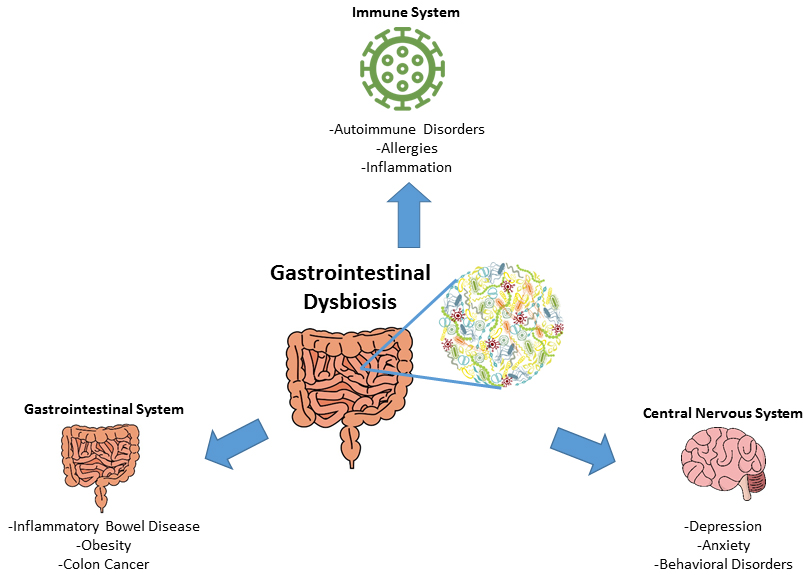
Figure 1. Gastrointestinal Dysbiosis. Various diseases of the gastrointestinal, central nervous and immune systems have been associated with gastrointestinal microbial dysbiosis (Figure designed by A. Buerger, from Buford 2017 and Kovatcheva-Datchary et al. 2013). |
The gastrointestinal microbiome performs many essential functions in the human body. First, it processes macronutrients for uptake and use in essential cellular processes throughout the body (Theriot et al. 2013; Kovatcheva-Datchary et al. 2013). Second, it produces signaling molecules capable of interacting with human receptors throughout the body, suggesting a role for the microbiome in the regulation of homeostasis (Cohen et al. 2017; Davison et al. 2017). Finally, it provides essential immune functions, protecting the body from infections and other immune responsive diseases (Burcelin 2016). These are just a few of the many critical functions that the gastrointestinal microbiome plays in maintaining host health.
The composition of a ’healthy’ microbiome (or gastrointestinal symbiosis) can vary individually, temporally, and geographically. Changes in the diversity and abundance of microbiota of an individual’s core microbiome are defined as microbial dysbiosis. Dysbiosis has been associated with the development of acute and chronic diseases, including inflammatory bowel diseases, metabolic disorders like obesity and diabetes, and psychological disorders like depression and anxiety (Figure 1). The etiology of gastrointestinal microbial dysbiosis (i.e. growth of harmful microbes, overgrowth of microorganism symbionts, disequilibrium of the bacterial community) was historically associated with lifestyle factors including diet and exercise. Recent evidence suggests that chemical exposures as well as antimicrobial agents can cause this physiological disturbance.
Antimicrobial agents are designed to eliminate and prevent microbial infections. An unintended consequence of their use, however, is the elimination of the ‘good’ gastrointestinal microbiota that have beneficial functions for the host (symbiotic). These changes can lead to gastrointestinal microbial dysbiosis. For example, triclosan is an antimicrobial chlorinated aromatic biocide that has been widely used in many personal care products (e.g. toothpaste, soap, cookware, furniture, etc.). Evidence has demonstrated that triclosan exposure can cause changes in microbiota abundance and pathogen interactions, making the host more susceptible to disease (Gaulke et al. 2016).
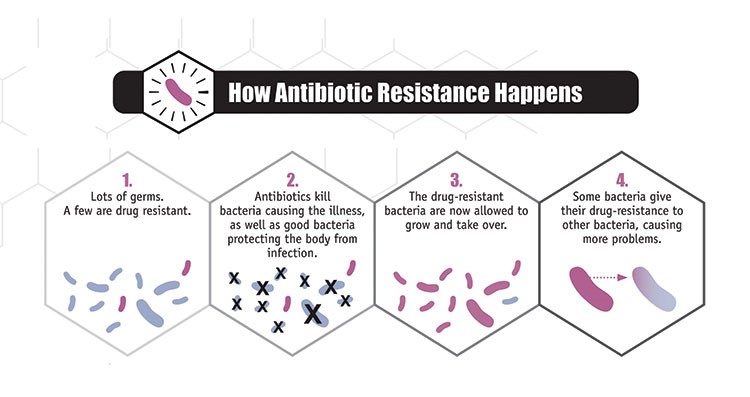
Figure 2. Antibiotic Resistance. Process by which antibiotic resistance in the gastrointestinal microbiome can occur through the following process (Source: CDC, https://www.cdc.gov/narms/faq.html). |
Furthermore, the widespread and indiscriminate use of antimicrobial agents can lead to the development of microbial resistant strains. For example, bacteria that are not affected by antibiotic treatment may survive and thrive, passing on resistance genes to other bacteria via horizontal gene transfer and subsequent generations (Sommer et al. 2009) (Figure 2). This can result in an increased susceptibility of the host to other pathogenic bacterial infections, since the elimination of competing species leaves an environment that is ripe for colonization (Theriot et al. 2013). The loss of essential bacteria during antibiotic treatment also decreases the production of metabolic products, which can significantly alter the availability of essential nutrients to the host (Theriot et al. 2013).
While antimicrobial agents are essential in reducing the incidence of specific infectious diseases in medical and public health interventions, indiscriminate and overuse of these compounds can negatively impact human health, as it relates to the gastrointestinal microbiome. They should be used as prescribed in efforts to minimize the risk of antimicrobial resistance and the development of microbiota-related illnesses. Utilizing the One Health concept, we can understand the importance of the vast gastrointestinal microbiota ecosystem as it relates to human health, examine the contributions of societal and environmental factors to antimicrobial usage, and promote their proper use in efforts to minimize the risk of antimicrobial resistance.
References:
-
Buford, T. W. (2017). (Dis)Trust your gut: the gut microbiome in age-related inflammation, health, and disease. Microbiome, 5(1), 80.
-
Burcelin, R. (2016). Gut microbiota and immune crosstalk in metabolic disease. Molecular Metabolism, 5(9), 771-781.
-
Cohen, L. J., Esterhazy, D., Kim, S. H., Lemetre, C., Aguilar, R. R., Gordon, E. A., ... & Chu, J. (2017). Commensal bacteria make GPCR ligands that mimic human signalling molecules. Nature, 549(7670), 48.
-
Davison, J. M., Lickwar, C. R., Song, L., Breton, G., Crawford, G. E., & Rawls, J. F. (2017). Microbiota regulate intestinal epithelial gene expression by suppressing the transcription factor Hepatocyte nuclear factor 4 alpha. Genome Research, gr-220111.
-
Gaulke, C. A., Barton, C. L., Proffitt, S., Tanguay, R. L., & Sharpton, T. J. (2016). Triclosan exposure is associated with rapid restructuring of the microbiome in adult zebrafish. PLoS One, 11(5), e0154632.
-
Kovatcheva-Datchary, P., Tremaroli, V., & Bäckhed, F. (2013). The gut microbiota. In E. Rosenberg, E. F. DeLong, S. Lory, E. Stackebrandt, F. Thompson (Ed.), The prokaryotes (pp. 3-24). Berlin, Heidelberg: Springer.
-
Sender, R., Fuchs, S., & Milo, R. (2016). Are we really vastly outnumbered? Revisiting the ratio of bacterial to host cells in humans. Cell, 164(3), 337-340.
-
Sommer, M. O., Dantas, G., & Church, G. M. (2009). Functional characterization of the antibiotic resistance reservoir in the human microflora. Science, 325(5944), 1128-1131.
-
Theriot, C. M., Koenigsknecht, M. J., Carlson Jr, P. E., Hatton, G. E., Nelson, A. M., Li, B., ... & Young, V. B. (2014). Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nature Communications, 5, 3114.
Next story: Review: Zoonotic Enteric Parasites: A One Health Guide to Preventing Infection
K-State Olathe
22201 W. Innovation Dr.
Olathe, KS 66061-1304
913-541-1220
913-541-1488 fax
olatheinfo@k-state.edu