Ruminant Microbiome as a Potential Reservoir of Antimicrobial Resistance: One Health Strategies for Reducing the Impact
Author
Tariku Jibat Beyene, DVM, MSc, PhD
Postdoctoral Research Associate
Center for Outcomes Research and Epidemiology (CORE)
College of Veterinary Medicine, Kansas State University
The emergence and spread of antimicrobial resistance (AMR) is a global issue, and it has become a major political, social, and economic problem of our time. Antimicrobial usage in livestock has been a major focus of this issue, because it has the potential to result in the direct spread of antimicrobial resistant bacteria to humans (Spellberg et al. 2016). With regard to AMR, projections suggest that by 2050, more people will die of bacterial infections than cancer due to the fact that currently available antimicrobials will no longer be as effective in treating bacterial infections. This will not only affect the health outcomes of humans, but it will also affect those of animals, including production yields of food-producing animals (O’Neill 2016). Thus, control and prevention of AMR requires the urgent adoption of a “One Health” approach, promoted through the integration of human, animal, and environmental health (Chainier et al. 2017; Oura et al. 2017). This article not only discusses ruminants and the potential source of antimicrobial resistant bacteria from the microbiota of their gastrointestinal tract, but also focuses on general strategies for reducing AMR across the globe.
Antimicrobial resistance and ruminants
Ruminants are a group of animals that are competitively advantageous in terms of resources, as they consume feedstuffs, including forages and by-products that cannot be used by non-ruminants, and they do not compete with humans for food (Auffret et al. 2017). Because of their unique digestive system with four stomach compartments, pre-gastric fermentative digestion of feed occurs in the rumen, which is a microbiological vat composed of both bacteria and protozoa that are responsible for digesting up to 70-80% of digestible dry matter.
In addition to therapeutic use, antimicrobials are often used in ruminant feed as additives to: 1) counter the ill effects of diets with rapidly digestible carbohydrates, 2) improve feed efficiency, and 3) reduce liver abscess incidences (Thomas et al. 2017). However, antimicrobials as feed additives can also lead to the selection of antimicrobial resistant bacteria in the rumen that express AMR genes (Cameron and McAllister 2016). Thus, increases in the abundance of bacteria with AMR genes in the gastrointestinal tract and feces of ruminant livestock have to potential to be a significant source of antimicrobial resistant bacteria to humans (Penders et al. 2013).
Antimicrobial resistance and the ruminant microbiome: The problem
Figure 1. Schematic Illustration of Possible Links Between Antibiotic Use in Animals and Human Disease. The prevalence of resistant bacteria in ruminants is influenced by antibiotic use in that setting. The impact of infection depends crucially on the capacity for sustained human-to-human (H2H) transmission. Arrows linking the two populations represent: a) direct transmission of bacteria not adapted to transmission in humans via the food chain (e.g., Campylobacter, Salmonella) or direct contact with animals; b) direct transmission of organisms already adapted to transmission in humans; c) transfer of resistance genes from ruminants into pathogens unlimited in their ability to be transmitted among humans. (Figure adapted from Chang et al. 2015). |
Humans are at risk for exposure to bacteria with AMR genes from animals through direct contact, ingestion of contaminated food or water, and/or contact with infected humans. As described by Chang et al. 2015, the prevalence of resistant bacteria in ruminants is influenced by antibiotic use in that setting, and the resultant impact of transmitting AMR bacteria from ruminants to humans depends critically on the capacity for sustained human-to-human (H2H) transmission (as shown in Figure 1). Three mechanisms have been described that could explain the means by which AMR in ruminants can lead to this threat in humans (Lipsitch et al. 2002):
i) A human becomes infected by a resistant microorganism of animal origin through either contact with livestock or through ingestion of contaminated meat or water without ongoing transmission of the pathogen between humans.
ii) A human becomes infected and/or colonized with a human-adapted resistant microorganism through any of the means described above, followed by ongoing transmission among humans, with some of these humans becoming ill. This scenario constitutes a break in the ‘species barrier’ by a microbe that may be directly pathogenic to humans or may be a commensal with the ability to cause opportunistic infections.
iii) A human becomes infected and/or colonized with a resistant microorganism through any of the means described above, and AMR genes arising from the ruminant gut bacteria are introduced to human pathogens by horizontal gene transfer. The resulting resistant lineages of bacteria are then further selected based on antibiotic use in humans (Chang et al. 2015).
The emergence and spread of AMR from agricultural sources can be exacerbated for many reasons, but are not limited to the following (Uchil et al. 2014):
i) Weak regulations of the production and sale of antibiotics and the absence of credible drug regulations and law enforcement, especially in developing countries,
ii) Indiscriminate use of antibiotics in livestock and aquaculture for treatment and growth promotion,
iii) Lack of system-wide monitoring of measures, and
iv) Inadequate research and development of new antimicrobials. There is a tremendous need to establish new treatments while preserving the efficacy of existing ones.
One Health approach: The solution
A holistic, One health approach of combating the problem of AMR provides a collaborative concept to broaden engagement beyond the realm of health and science. An interconnected and integrated One Health surveillance framework that puts at its center both antimicrobial consumption and AMR would benefit in combating the problem (Queenan et al. 2016). Like the concept of One Health, this significant problem in terms of engagement requires an approach from different perspectives and levels of authority to become successful with positive outcomes. For instance, in Europe, when the use of the antibiotic, avoparcin, in animal feed was determined to be an important driver of the development of vancomycin-resistant enterococci (VRE) and consequent colonization in the human intestine, its use in animal feed was banned, resulting in a dramatic reduction of human cases with VRE (Marshall and Levy 2011).
Currently, surveillance is considered a major asset for programs directed toward AMR. The objective of surveillance programs is to facilitate the containment of antibiotic resistance. It is a useful tool that generates data on antimicrobial use and AMR, which is essential in updating national essential drug lists (EDLs) and formulating infection control policies. It may also be helpful in improving the prescription of antimicrobials and the development of empirical therapies or improved standards of treatment.
Additionally, besides effective legislation and surveillance programs to decrease human exposure to AMR bacteria from livestock, there are additional practices beginning at the farm-level that could further reduce the use of antimicrobials in animals and potential exposure to humans, including improved sanitation practices, provision of probiotics or nutritional supplements in animal feed, and vaccination for common animal diseases.
|
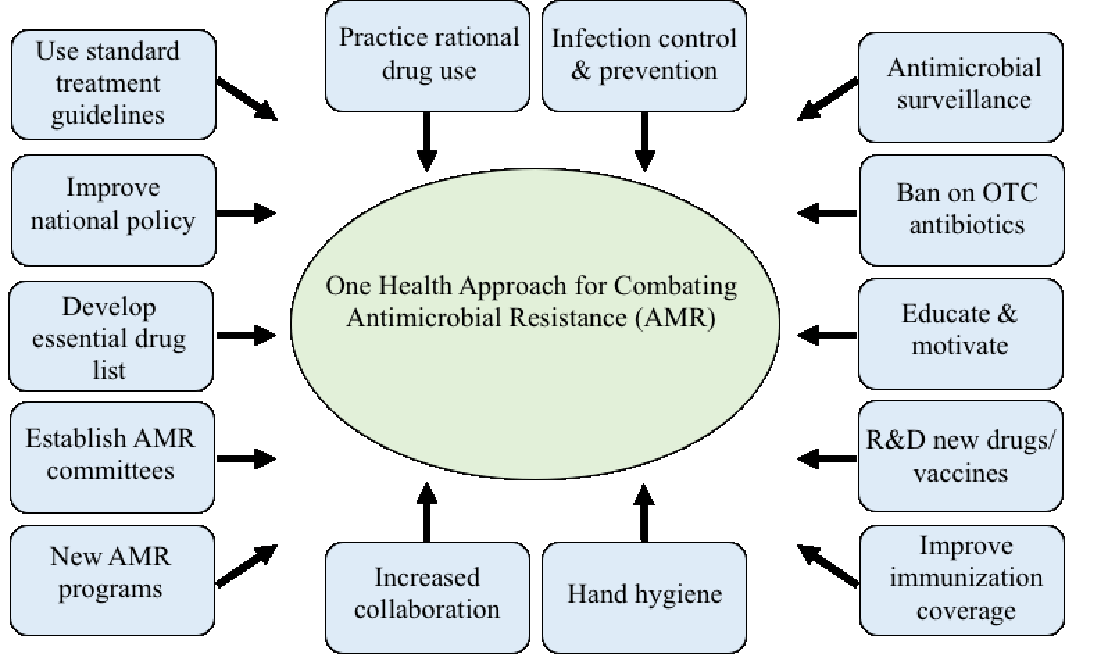
According to Uchil et al. (2014), potential measures to further combat AMR include international measures, national strategies, and actions at both the community and local levels (Figure 2). Some strategies using this multifaceted approach are listed below:
i) International measures, as recommended by the World Health Organization (WHO), would include:
-
- Increasing collaborations between governments, nongovernmental organizations, professional groups, and international agencies,
- Developing new networks that undertake surveillance of antimicrobial use and AMR,
- Promoting an international approach for control of counterfeit antimicrobials,
- Incentivizing research and development of new drugs and vaccines, and
- Forming new and reinforcing existing programs to contain AMR.
ii) National strategies would include the development of national committees with intersectoral coordination and regulatory actions.
iii) Action at the community level would include rational use of prescribed and over-the-counter antibiotics, developing guidelines for use of antibiotics at local levels, and instilling the practice of good standard hygiene.
iv) In local healthcare facilities, recommended measures should be followed to prevent and control infection, including the establishment of infection prevention and control (IPC) committees, good hand hygiene practices, accurate diagnosis and treatment of infections, rational antimicrobial use, and surveillance and reporting of AMR and antibiotic use within health facilities.
In conclusion, One Health strategies for reducing the impact of AMR should begin by considering all potential sources of antimicrobial resistant bacteria, including how routine practices in farming communities with food-producing animals influence the development and potential spread of AMR to humans. An interconnected and integrated One Health surveillance framework may provide effective input on best practices when it comes to antimicrobial use for treatment. Lastly, since AMR is a global issue, it will require coordination, cooperation, and action that spans from local healthcare communities to the international stage in order to make a difference in the health outcomes of humans and animals in the not too distant future.
References:
Auffret, M. D., Dewhurst, R. J., Duthie, C.-A., Rooke, J. A., Wallace, R. J., Freeman, T. C.,… Roehe, R. (2017). The rumen microbiome as a reservoir of antimicrobial resistance and pathogenicity genes is directly affected by diet in beef cattle. Microbiome, 5(1), 159.
Cameron, A., & McAllister, T. A. (2016). Antimicrobial usage and resistance in beef production. J Anim Sci Biotechnol, 7(1), 68.
Chainier, D., Barraud, O., Masson, G., Couve-Deacon, E., François, B., Couquet, C.-Y., & Ploy, M.-C. (2017). Integron digestive carriage in human and cattle: A “One Health” cultivation-independent approach. Front Microbiol, 8, 1891.
Chang, Q., Wang, W., Regev‐Yochay, G., Lipsitch, M., & Hanage, W. P. (2015). Antibiotics in agriculture and the risk to human health: how worried should we be? Evol Appl, 8(3), 240-247.
Lipsitch, M., Singer, R. S., & Levin, B. R. (2002). Antibiotics in agriculture: When is it time to close the barn door? Proc Natl Acad Sci, 99(9), 5752-5754.
Marshall, B. M., & Levy, S. B. (2011). Food animals and antimicrobials: impacts on human health. Clin Microbiol Rev, 24(4), 718-733.
O’Neill, J. (2016). Review on antimicrobial resistance: tackling drug-resistant infections globally—final report and recommendations (Wellcome Trust, UK Government, 2016).
Oura, C., Mahase-Gibson, A., & Stephen, C. (2017). Caribbean resilience and prosperity through One Health. The University of West Indies St. Augustine, Trinidad & Tobago. http://www.cwhc-rcsf.ca/docs/technical_reports/Caribbean_Resilience.pdf
Penders, J., Stobberingh, E. E., Savelkoul, P. H., & Wolffs, P. (2013). The human microbiome as a reservoir of antimicrobial resistance. Front Microbiol, 4, 87.
Queenan, K., Häsler, B., & Rushton, J. (2016). A One Health approach to antimicrobial resistance surveillance: is there a business case for it? Int J Antimicrob Agents, 48(4), 422-427.
Spellberg, B., Hansen, G. R., Kar, A., Cordova, C. D., Price, L. B., & Johnson, J. R. (2016). Antibiotic resistance in humans and animals. Discussion Paper, National Academy of Medicine, Washington, DC. http://www.nam.edu/antibiotic-resistance-in-humans-and-animals
Thomas, M., Webb, M., Ghimire, S., Blair, A., Olson, K., Fenske, G. J., . . . Scaria, J. (2017). Metagenomic characterization of the effect of feed additives on the gut microbiome and antibiotic resistome of feedlot cattle. Sci Rep, 7(1), 12257.
Uchil, R. R., Kohli, G. S., KateKhaye, V. M., & Swami, O. C. (2014). Strategies to combat antimicrobial resistance. J Clin Diagn Res, 8(7), ME01-4.
Next story: Outsmarting antibiotic resistance is topic of Kansas City One Health Day on Nov. 1
Contact Us